Preface
Goal: A practical case to collect unique record fields using GNU.
GNU R has different beauty compared with Julia.
We can simply utilize dataframe to data handle records.
We have to switch our paradigm to dataframe,
instead of general coding approach.
Hence, a closer to statistical approach,
in which GNU R is made for.
It is actually just R, without GNU.
I try to use R-lang, but it could be mistakenly referred as erlang.
Reference Reading
Source Examples
You can obtain source examples here:
Common Use Case
Task: Get the unique tag string
Please read overview for more detail.
Prepopulated Data
Songs and Poetry
library(tibble)
tibble(
title = "Cantaloupe Island",
tags = list(list("60s", "jazz"))
) %>% add_row(
title = "Let It Be",
tags = list(list("60s", "rock"))
) %>% add_row(
title = "Knockin' on Heaven's Door",
tags = list(list("70s", "rock"))
) %>% add_row(
title = "Emotion",
tags = list(list("70s", "pop"))
) %>% add_row(
title = "The River"
) -> dataframeGNU R Solution
The Answer
There might be many ways to do things in GNU R.
One of them is this oneliner as below:
library(tibble)
library(purrr)
load("songs.RData")
(dataframe %>%
dplyr::filter(.data$tags != "NULL")
)$tags %>% flatten %>% unique %>% pasteEnough with introduction, at this point we should go straight to coding.
Environment
No need any special setup. Just run and voila..!
You can utilize R-Studio.
But for this simple case in this article,
any text editor sufficient.
An optional packages called purrr,
need to be setup manually.
1: Data Structure
We are going to use list throught out this article.
Simple List
Before building a struct,
I begin with simple array.
tags <- list("rock", "jazz", "rock", "pop", "pop")
paste(tags)It is easy to dump variable in GNU R using paste.
With the result similar as below string:
❯ Rscript 01-tags.r
[1] "rock" "jazz" "rock" "pop" "pop" While print show the complete structural data,
paste is actually just concatenation of the data interpretation.
The Record Structure
We can build record structure using dataframe.
The convenient way to add record like data is by using tibble.
First we setup two columns title and tags, shown in below code:
library(tibble)
dataframe <- tibble(
title = "Cantaloupe Island",
tags = list(list("60s", "jazz"))
)
print(dataframe)With the result similar as below dataframe:
$ Rscript 02-dataframe.r
# A tibble: 1 x 2
title tags
<chr> <list>
1 Cantaloupe Island <list [2]>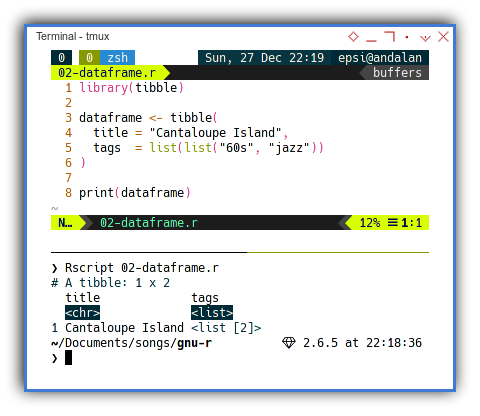
We can explore more about dataframe.
paste(head(dataframe))
paste(names(dataframe))
paste(dataframe["tags"])
paste(dataframe$tags)With the result similar as below string:
[1] "Cantaloupe Island" "list(list(\"60s\", \"jazz\"))"
[1] "title" "tags"
[1] "list(list(\"60s\", \"jazz\"))"
[1] "list(\"60s\", \"jazz\")"Issue with List in Rows
Why double list 🤔?
You might spot a strange declaration here:
dataframe <- tibble(
title = "Cantaloupe Island",
tags = list(list("60s", "jazz"))
)This double list is a workaround. If we declare as below:
dataframe <- tibble(
title = "Cantaloupe Island",
tags = list("60s", "jazz")
)The list will be expanded into two rows of data.
Rows of Song Struct
Meet The Songs Rows
From just karaoke, we can go pro with recording studio. From simple data, we can build a structure to solve our task.
As has been said,
the convenient way to add-row is by using tibble.
library(tibble)
dataframe <- tibble(
title = "Cantaloupe Island",
tags = list(list("60s", "jazz"))
)
dataframe <- add_row(dataframe,
title = "Let It Be",
tags = list(list("60s", "rock"))
)
dataframe <- dataframe %>% add_row(
title = "Knockin' on Heaven's Door",
tags = list(list("70s", "rock"))
)
add_row(dataframe,
title = "Emotion",
tags = list(list("70s", "pop"))
) -> dataframe
dataframe %>% add_row(
title = "The River"
) -> dataframe
dataframe %>% printWith the result similar as below dataframe:
$ Rscript 03-add-row.r
# A tibble: 5 x 2
title tags
<chr> <list>
1 Cantaloupe Island <list [2]>
2 Let It Be <list [2]>
3 Knockin' on Heaven's Door <list [2]>
4 Emotion <list [2]>
5 The River <NULL> You can spot that there are four ways to manipulate data.
- Two combinations of assignment
<-operator, and. - Two combinations of pipe
%>%operator, and.
Maybe Null
We need, a not too simple, case example.
GNU R handle this NULL value natively in statistical context.
I won’t bother into the the detail.
Let’s just the GNU R handle this with its approach.
2: Separating Module
Since we need to reuse the songs rows multiple times, it is a good idea to separate the record from logic.
Songs Module
The code can be shown as below:
library(tibble)
tibble(
title = "Cantaloupe Island",
tags = list(list("60s", "jazz"))
) %>% add_row(
title = "Let It Be",
tags = list(list("60s", "rock"))
) %>% add_row(
title = "Knockin' on Heaven's Door",
tags = list(list("70s", "rock"))
) %>% add_row(
title = "Emotion",
tags = list(list("70s", "pop"))
) %>% add_row(
title = "The River"
) -> dataframe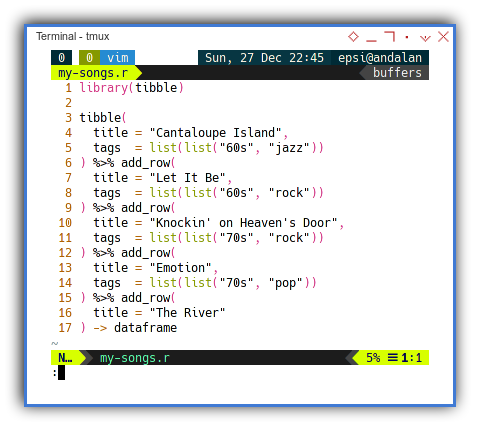
The pipe %> syntatic sugar from tibble is very nice.
Using Songs Module
Now we can have a very short code.
source("my-songs.r")
dataframe %>% printWith the result exactly the same as above array.
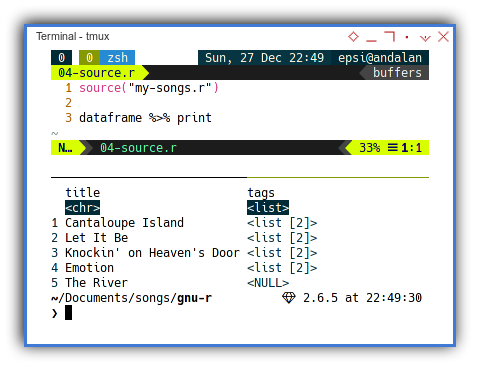
Julia has this map do notation.
3: External Data
RData and CSV
By its design, GNU R can handle large amount of external data.
Why don’t we try it now.
RData: Write
We can save the state of the object into a file.
source("my-songs.r")
save(dataframe, file = "songs.RData")This way, current object can be modularized into a file, to be used later, in other script.
RData: Read
This is how we read it in other script.
library(tibble)
load("songs.RData")
dataframe %>% printThe object name persist. No need any new assignment.
We can continue from where we left.
CSV: Write
There is another way,m using CSV. Bu unfortunately we use list with already has a comma separated value. So we cannot save into regular CSV.
A workaround can be done, to save this using CSV2.
This csv2 utilize ; as separator.
The list in each itself is also an issue.
A workaround is to conert to matrix first.
source("my-songs.r")
matrix <- as.matrix(dataframe)
matrix %>% print
matrix %>% write.csv2("./songs.csv")The matrix transformation has form as below:
$ Rscript 05-write-csv.r
title tags
[1,] "Cantaloupe Island" List,2
[2,] "Let It Be" List,2
[3,] "Knockin' on Heaven's Door" List,2
[4,] "Emotion" List,2
[5,] "The River" NULL CSV: Output
The CSV output result can be show as below:
"";"title";"tags"
"1";Cantaloupe Island;list("60s", "jazz")
"2";Let It Be;list("60s", "rock")
"3";Knockin' on Heaven's Door;list("70s", "rock")
"4";Emotion;list("70s", "pop")
"5";The River;NULLCSV: Read
We can read the CSV using code below.
library(tibble)
songs_df <- read.csv2(file = "./songs.csv")
songs_df["tags"] %>% pasteCSV: Caveat
CSV is good for plain records,
especially for data interchange between application.
But it is not recommended for storing list object.
Consider have a look at the output of the read below:
[1] "1:5"
[2] "c(\"Cantaloupe Island\", \"Let It Be\", \"Knockin' on Heaven's Door\", \"Emotion\", \"The River\")"
[3] "c(\"list(60s, jazz)\", \"list(60s, rock)\", \"list(70s, rock)\", \"list(70s, pop)\", \"NULL\")" What missing here is, the tags rows stored as string instead of list.
This become troublesome for further processing.
4: Extracting Fields
Walk, Filter, Drop/Keep
Walk
Map
Sometimes, we need to examine each records.
Not as a whole dataframe, but each rows separately.
To achieve this we can utilizr walk function from purrr library.
source("my-songs.r")
library(purrr)
dataframe$tags %>%
walk(function(current) {
current %>% as.list %>% paste %>% print
})With the result similar as below sequential lines of row:
$ Rscript 07-purrr.r
[1] "60s" "jazz"
[1] "60s" "rock"
[1] "70s" "rock"
[1] "70s" "pop"
character(0)Filter
To get rid of the row without tags data, we utilize dplyr::filter.
source("my-songs.r")
songs_df <- dataframe %>%
dplyr::filter(.data$tags != "NULL")
songs_df %>% printWith the result similar as below dataframe:
$ Rscript 08-filter.r
# A tibble: 4 x 2
title tags
<chr> <list>
1 Cantaloupe Island <list [2]>
2 Let It Be <list [2]>
3 Knockin' on Heaven's Door <list [2]>
4 Emotion <list [2]>Just beware of the fully qualified name,
or we may end up using filter from other library.
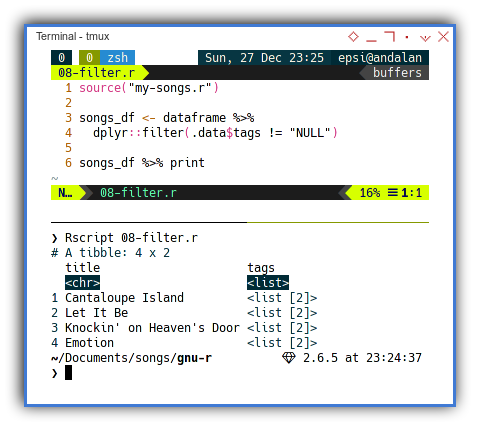
Drop/Keep
Although not required, we can shrink the dataframe,
to have only specific columns.
library(tibble)
songs_df <- read.csv2(file = "./songs.csv")
songs_df <- songs_df %>%
dplyr::filter(.data$tags != "NULL")
keeps <- "tags"
tags_df <- songs_df[ , keeps, drop = FALSE]
tags_df %>% printWith the result similar as below lines of string, loaded from csv.
$ Rscript 09-keep.r
tags
1 list(60s, jazz)
2 list(60s, rock)
3 list(70s, rock)
4 list(70s, pop)5: Finishing The Task
Flatten, Unique, Oneliner Pipe
Flatten
There is already a flatten function in dplyr library,
source("my-songs.r")
library(purrr)
songs_df <- dataframe %>%
dplyr::filter(.data$tags != "NULL")
tags_df <- songs_df[ , "tags", drop = FALSE]
flattened <- flatten(tags_df$tags)
flattened %>% pasteWith the result similar as below string:
$ Rscript 10-flatten.r
[1] "60s" "jazz" "60s" "rock" "70s" "rock" "70s" "pop" We don’t really need to drop columns. We can safely remove this line:
tags_df <- songs_df[ , "tags", drop = FALSE]Unique
There is also a unique function in standard library.
So I guess our problem is solved completely.
source("my-songs.r")
library(purrr)
songs_df <- dataframe %>%
dplyr::filter(.data$tags != "NULL")
flattened <- flatten(songs_df$tags)
distinct <- unique(flattened)
distinct %>% paste$ Rscript 11-unique.r
[1] "60s" "jazz" "rock" "70s" "pop" Oneliner
Clean up
Using a bunch of %> pipe operator,
we can completely transform code above,
into a single oneliner statement.
library(tibble)
library(purrr)
load("songs.RData")
(dataframe %>%
dplyr::filter(.data$tags != "NULL")
)$tags %>% flatten %>% unique %>% pasteThe code is now simple, and clear. We can understand exactly what it does, by just reading the code.
What is Next 🤔?
Consider continue reading [ Nim - Playing with Records ].